C2M2
Table of Contents
- Introduction
- Resources
- Frictionless Data Packages
- Identifiers
- C2M2 Tables
- Reference Tables
- Submission Guide
- Tutorial
Introduction
Schematic of C2M2
(please right-click on the image to see options to view a larger image in a different tab)
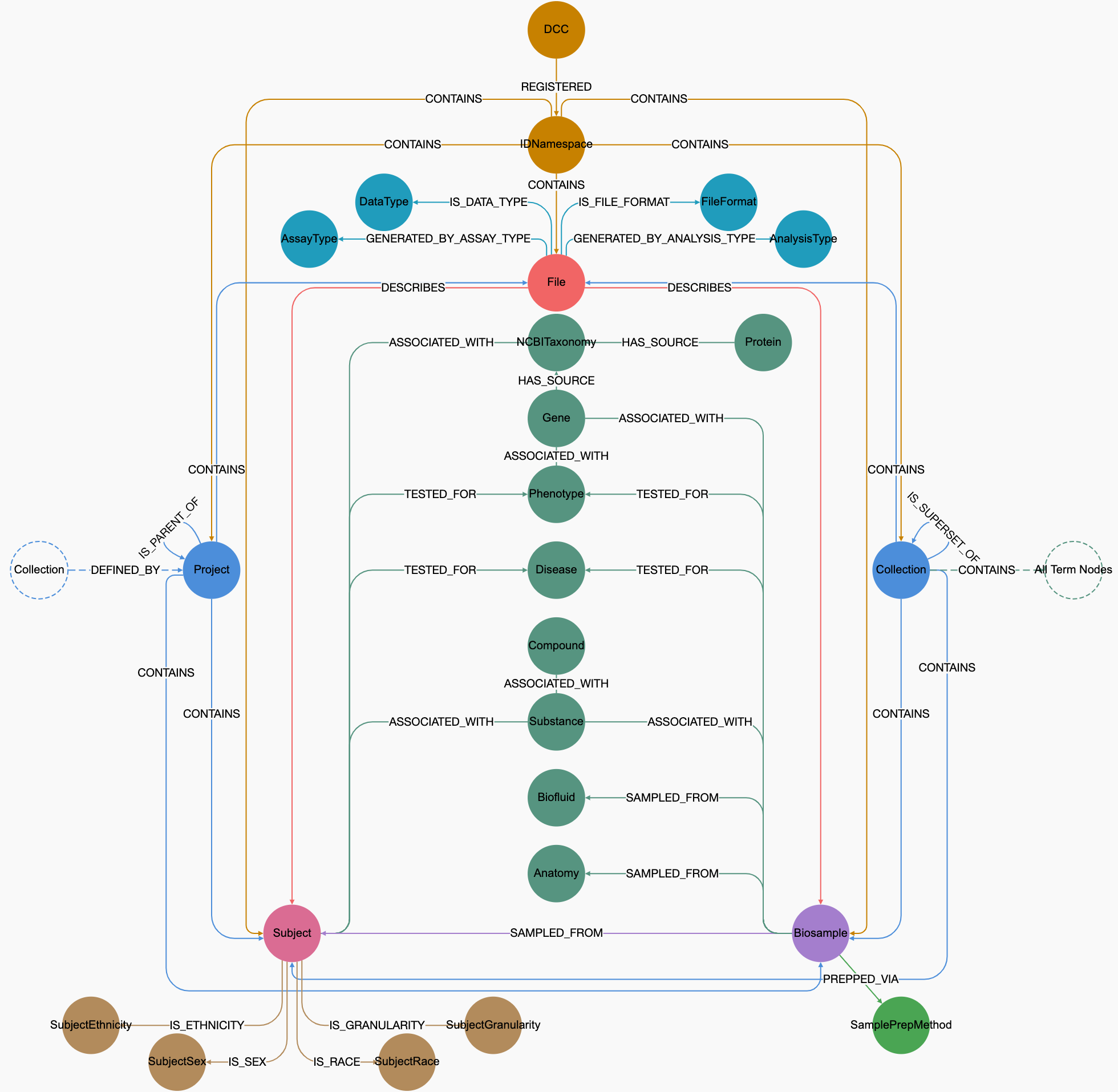
The Crosscut Metadata Model (C2M2) is a flexible metadata standard for describing experimental resources in biomedicine and related fields. A complete C2M2 submission, also called an "instance" or a "datapackage", is a zipped folder containing multiple tab-delimited files (TSVs) representing metadata records along with a JSON schema. To read more about datapackages, skip to the Frictionless Data Packages section.
Each TSV file is a data table containing various data records (rows) and their values for different fields (columns). Entity tables describe various types of data objects, while association tables describe the relationships between different entities.
Resources
The cfde-c2m2 repository includes a CLI helper for preparing, validating, and submitting your C2M2 submission.
Frictionless Data Packages
C2M2 instances are also known as "datapackages" based on the Data Package meta-specification from Frictionless Data.
From the original C2M2 documentation:
The Data Package meta-specification is a platform-agnostic toolkit for defining format and content requirements for files so that automatic validation can be performed on those files, just as a database management system stores definitions for database tables and automatically validates incoming data based on those definitions. Using this toolkit, the C2M2 JSON Schema specification defines foreign-key relationships between metadata fields (TSV columns), rules governing missing data, required content types and formats for particular fields, and other similar database management constraints. These architectural rules help to guarantee the internal structural integrity of each C2M2 submission, while also serving as a baseline standard to create compatibility across multiple submissions received from different DCCs.
Identifiers
In order to standardize and integrate information across DCCs, there must be a system of assigning unambiguous identifiers to individual DCC concepts and resources. These are the "C2M2 IDs", consisting of a id_namespace prefix and local_id suffix. Additionally, the C2M2 also allows individual resources to be assigned a persistent_id.
From the original C2M2 documentation:
Optional
persistent_ididentifiers are meant to be stable enough to be scientifically cited, and to provide for further investigation by accessing related resolver services. To be used as a C2M2persistent_id, an ID
- will represent an explicit commitment by the managing DCC that the attachment of the ID to the resource it represents is permanent and final
- must be a format-compliant URI or a compact identifier, where the protocol (the "scheme" or "prefix") specified in the ID is registered with at least one of the following (see the given lists for examples of URIs and compact identifiers)
- the IANA (list of registered schemes)
- scheme used must be assigned either "Permanent" or "Provisional" status
- Identifiers.org (list of registered prefixes)
- N2T (Name-To-Thing) (list of registered prefixes)
- protocols not appearing in the above registries but explicitly approved by the CFDE-CC. Currently, this list is limited to one protocol, namely drs:// URIs identifying GA4GH Data Repository Service resources.
- if representing a file, an ID used as a
persistent_idcannot be a direct-download URL for that file: it must instead be an identifier permanently attached to the file and only indirectly resolvable (through the scheme or prefix specified within the ID) to the file itself
C2M2 Tables
Sourced from the CFDE-CC Documentation Wiki
All of the tables in a C2M2 datapackage are inter-linked via foreign key relationships, as shown in the following diagram of the complete C2M2 system.
Entity relationship diagram of C2M2
(please right-click on the image to see options to view a larger image in a different tab)
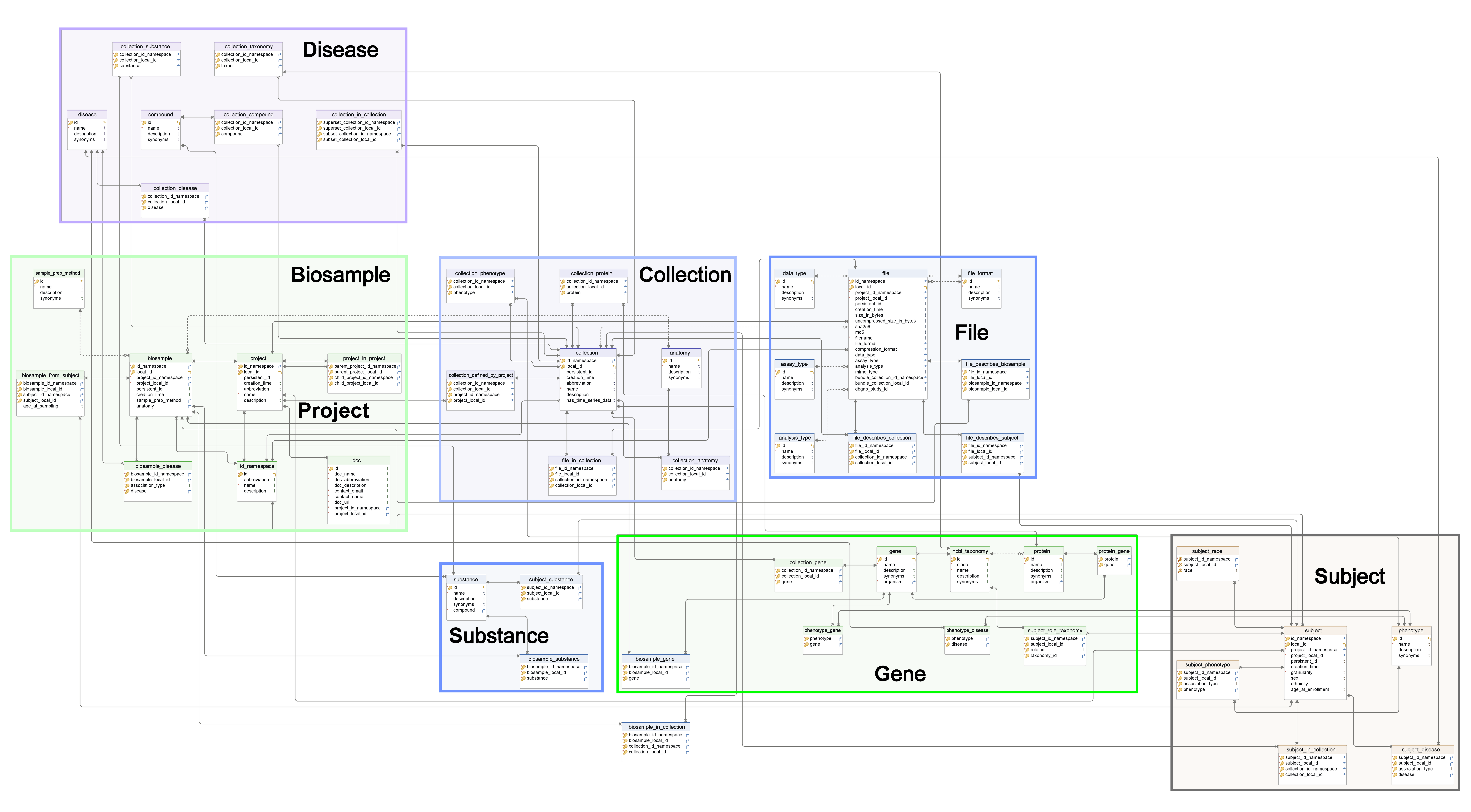
Crosscut Metadata Model (C2M2) Common Vocabulary (CV) Tables
These files can be assembled mostly automatically, please see the Submission Guide for instructions on assembling these files.
- All table files listed in this summary will be bundled together, along with the C2M2 datapackage JSON Schema file which defines them, to create a valid C2M2 datapackage for submission to CFDE
- TSV files for any empty (unused) tables must still be submitted, with only the (tab-separated) column-header row filled in
- Table (TSV) filenames must exactly match those listed in the JSON Schema file (and in these docs)
- Table column headers must exactly match those listed in the JSON Schema file (and in these docs)
- Table columns must appear in the order given in the JSON Schema file (and in these docs)
- Tables marked "CV term table" will be built automatically with the CFDE tools (wiki)
- Table (TSV) files must not contain any empty rows or extra lines
- Every TSV file must end with the final row of table data, terminated by a newline
| Table (click for detailed information) | Construction | Can be empty? | Notes |
|---|---|---|---|
| analysis_type.tsv | Built by script | Y | CV term table |
| anatomy.tsv | Built by script | Y | CV term table |
| assay_type.tsv | Built by script | Y | CV term table |
| biofluid.tsv | Built by script | Y | CV term table |
| biosample.tsv | Prepared by submitter | Y | This table will have one row for each biosample |
| biosample_disease.tsv | Prepared by submitter | Y | For biosamples with disease metadata, this table will have one row for each disease associated with each biosample, along with a field distinguishing "exemplar of disease" from "disease specifically ruled out" |
| biosample_from_subject.tsv | Prepared by submitter | Y | This table will have one row for each attribution of a biosample to a subject |
| biosample_gene.tsv | Prepared by submitter | Y | For each biosample with a small group of associated genes (e.g. knockdown targets), this table will have one row for each association of a gene with a biosample |
| biosample_in_collection.tsv | Prepared by submitter | Y | This table will have one row for each assignment of a biosample as a member of a collection |
| biosample_ptm.tsv | Prepared by submitter | Y | For each biosample with a small group of associated PTMs, this table will have one row for each association of a PTM with a biosample |
| biosample_substance.tsv | Prepared by submitter | Y | For biosamples with substance metadata, this table will have one row for each association of a substance with a biosample |
| collection.tsv | Prepared by submitter | Y | This table will have one row for each collection |
| collection_anatomy.tsv | Prepared by submitter | Y | Each row in this table is equivalent to the statement "the contents of collection X directly relate to the study of anatomy Y", for one particular (collection X, anatomy Y) pair |
| collection_biofluid.tsv | Prepared by submitter | Y | Each row in this table is equivalent to the statement "the contents of collection X directly relate to the study of biofluid Y", for one particular (collection X, biofluid Y) pair |
| collection_compound.tsv | Prepared by submitter | Y | Each row in this table is equivalent to the statement "the contents of collection X directly relate to the study of compound Y", for one particular (collection X, compound Y) pair |
| collection_defined_by_project.tsv | Prepared by submitter | Y | This table will have one row for each collection that was generated directly by a project listed in the project.tsv table |
| collection_disease.tsv | Prepared by submitter | Y | Each row in this table is equivalent to the statement "the contents of collection X directly relate to the study of disease Y", for one particular (collection X, disease Y) pair |
| collection_gene.tsv | Prepared by submitter | Y | Each row in this table is equivalent to the statement "the contents of collection X directly relate to the study of gene Y", for one particular (collection X, gene Y) pair |
| collection_in_collection.tsv | Prepared by submitter | Y | This table will have one row for each parent->child (collection->subcollection) relationship |
| collection_phenotype.tsv | Prepared by submitter | Y | Each row in this table is equivalent to the statement "the contents of collection X directly relate to the study of phenotype Y", for one particular (collection X, phenotype Y) pair |
| collection_protein.tsv | Prepared by submitter | Y | Each row in this table is equivalent to the statement "the contents of collection X directly relate to the study of protein Y", for one particular (collection X, protein Y) pair |
| collection_ptm.tsv | Prepared by submitter | Y | Each row in this table is equivalent to the statement "the contents of collection X directly relate to the study of PTM Y", for one particular (collection X, PTM Y) pair |
| collection_substance.tsv | Prepared by submitter | Y | Each row in this table is equivalent to the statement "the contents of collection X directly relate to the study of substance Y", for one particular (collection X, substance Y) pair |
| collection_taxonomy.tsv | Prepared by submitter | Y | Each row in this table is equivalent to the statement "the contents of collection X directly relate to the study of taxonomy Y", for one particular (collection X, taxonomy Y) pair |
| compound.tsv | Built by script | Y | CV term table |
| data_type.tsv | Built by script | Y | CV term table |
dcc.tsv (formerly primary_dcc_contact.tsv) | Prepared by submitter | N | This table will have exactly one row |
| disease.tsv | Built by script | Y | CV term table |
| domain_location.tsv | Prepared by submitter | Y | This table will have one row for each unique domain_location term in the ptm table |
| file.tsv | Prepared by submitter | Y | This table will have one row for each file |
| file_describes_biosample.tsv | Prepared by submitter | Y | This table will have one row for each association of a biosample with a describing file |
| file_describes_collection.tsv | Prepared by submitter | Y | This table will have one row for each association of a collection with a describing file |
| file_describes_subject.tsv | Prepared by submitter | Y | This table will have one row for each association of a subject with a describing file |
| file_format.tsv | Built by script | Y | CV term table |
| file_in_collection.tsv | Prepared by submitter | Y | This table will have one row for each assignment of a file as a member of a collection |
| gene.tsv | Built by script | Y | CV term table |
| id_namespace.tsv | Prepared by submitter | N | This table will have one row for each C2M2 identifier namespace registered with CFDE |
| ncbi_taxonomy.tsv | Built by script | Y | CV term table |
| phenotype.tsv | Built by script | Y | CV term table |
| phenotype_disease.tsv | Built by script | Y | Each row in this table is equivalent to the statement "phenotype X is known to be associated with disease Y", for one particular (phenotype X, disease Y) pair; contents are autoloaded from HPO by the submission prep script, which will add relevant rows for every phenotype term and every disease term used in submitter-prepared tables |
| phenotype_gene.tsv | Built by script | Y | Each row in this table is equivalent to the statement "phenotype X is known to be associated with gene Y", for one particular (phenotype X, gene Y) pair; contents are autoloaded from HPO by the submission prep script, which will add relevant rows for every phenotype term and every gene term used in submitter-prepared tables |
| project.tsv | Prepared by submitter | N | This table will have one row for each project |
| project_in_project.tsv | Prepared by submitter | Y* | This table will have one row for each parent->child (project->subproject) relationship. --- *If you have more than one project in your project.tsv table, then you must populate this table with all of your program's top-level projects, listed as children of your program's root project. |
| protein.tsv | Built by script | Y | CV term table |
| protein_gene.tsv | Built by script | Y | Each row in this table is equivalent to the statement "protein X is known to be associated with gene Y", for one particular (protein X, gene Y) pair; contents are autoloaded from HPO by the submission prep script, which will add relevant rows for every protein term and every gene term used in submitter-prepared tables |
| ptm.tsv | Prepared by submitter | Y | This table will have one row for each PTM |
| ptm_type.tsv | Prepared by submitter | Y | This table will have one row for each unique ptm_type term in the ptm table |
| ptm_subtype.tsv | Prepared by submitter | Y | This table will have one row for each unique ptm_subtype term in the ptm table |
| sample_prep_method.tsv | Built by script | Y | CV term table |
| subject.tsv | Prepared by submitter | Y | This table will have one row for each subject |
| subject_disease.tsv | Prepared by submitter | Y | For subjects with disease metadata, this table will have one row for each disease associated with each subject, along with a field distinguishing "disease detected" from "disease specifically ruled out" |
| subject_in_collection.tsv | Prepared by submitter | Y | This table will have one row for each assignment of a subject as a member of a collection |
| subject_phenotype.tsv | Prepared by submitter | Y | For every subject with phenotype metadata, this table will have one row for each phenotype associated with each subject, along with a field distinguishing "exemplar of phenotype" from "phenotype specifically ruled out" |
| subject_race.tsv | Prepared by submitter | Y | This table will have one row for each subject with a race assertion |
| subject_role_taxonomy.tsv | Prepared by submitter | Y | This table will have one row for each taxon assigned to a subject |
| subject_substance.tsv | Prepared by submitter | Y | For subjects with substance metadata, this table will have one row for each substance associated with each subject |
| substance.tsv | Built by script | Y | CV term table |
Reference Tables
Certain table fields within the C2M2 are restricted to only a few pre-defined values, such as biosample_disease.association_type or subject.granularity. Reference tables containing the allowed values for these fields, which were originally published on the CFDE-CC Documentation Wiki, are linked below:
Submission Guide
Sourced from the CFDE C2M2 Documentation
Step 1: Install CFDE C2M2 CLI
pipx install cfde-c2m2
Step 2: Initialize Submission
Initializes a fresh C2M2 submission in the current working directory, creating blank versions of all tables defined in the current C2M2 datapackage schema (C2M2_datapackage.json).
mkdir submission
cd submission
cfde-c2m2 init
Step 3: Fill in tables
Usually you would develop a script to format the information you already have into the structure of the C2M2 tables. The schema is designed to support incremental improvements and as such many tables and fields are optional. An example of doing this is available below in the tutorial.
Step 4: Prepare submission
Resolves ontology identifiers and populates the Common Vocabulary (CV) tables.
cfde-c2m2 prepare
Step 5: Validate
Checks validity of C2M2 submission and highlights tables for which corrections, if any, are needed.
cfde-c2m2 validate
Step 6: Package
Collects metadata tables in a zip file to be submitted to the DRC portal.
cfde-c2m2 package
Step 7: Submit
Currently, the CFDE Workbench Data Portal accepts complete datapackage submissions in ZIP file format (*.zip).
For specific instructions on using the submission system, see the Contribution Guide.
To submit a datapackage, navigate to the Submission System.
Tutorial
For the April 2022 CFDE Cross-Pollination meeting, the LINCS DCC demonstrated a Jupyter Notebook tutorial on building the file, biosample, and subject tables for LINCS L1000 signature data. The code and files can be found at the following link:
LINCS C2M2 Demo (04-05-2022 Cross-Pollination)
Code snippets from this tutorial corresponding to each step are reproduced below. Note that the C2M2 datapackage building process will vary across DCCs, depending on the types of generated data, ontologies, and access standards in place. In general, the process will be as follows:
-
Become familiar with the current structure of the C2M2, including the required fields across the entity and association tables, start with the steps from the submission guide:
# install the cfde-c2m2 helper CLI pipx install cfde-c2m2 # create a directory for the submission mkdir submission cd submission # initialize the submission directory, fetching the C2M2_datapackage.json and building blank tables cfde-c2m2 init -
Identify the relevant namespace for all files, and build the
id_namespaceanddcctables first.- For LINCS, the namespace "https://lincsproject.org" is used. The following code will generate the LINCS
id_namespacetable:
import pandas as pd pd.DataFrame([{ 'id': 'https://www.lincsproject.org', 'abbreviation': 'LINCS', 'name': 'Library of Integrated Network-Based Cellular Signatures', 'description': 'A network-based understanding of biology', }], columns=['id', 'abbreviation', 'name', 'description'] ).to_csv('id_namespace.tsv', sep='\t', index=False) - For LINCS, the namespace "https://lincsproject.org" is used. The following code will generate the LINCS
-
Identify all relevant projects and their associated files that will be included in the C2M2 datapackage. Generate container entity tables (
project,collection) that describe logic, theme, or funding-based groups of the core entities. Note that projects and collections may be nested, in theproject_in_projectandcollection_in_collectiontables.- In the LINCS tutorial, the data comes from the 2021 release of the LINCS L1000 Connectivity Map dataset. In creating a project representing the files from this dataset, there must also be an overarching root project for all LINCS data.
import pandas as pd from datetime import datetime pd.DataFrame([ { 'id_namespace': 'https://www.lincsproject.org', 'local_id': 'LINCS', 'persistent_id': 'https://www.lincsproject.org', 'creation_time': datetime(2013, 1, 1).astimezone().isoformat(), 'abbreviation': 'LINCS', 'name': 'Library of Integrated Network-Based Cellular Signatures', 'description': 'A network-based understanding of biology', }, { 'id_namespace': 'https://www.lincsproject.org', 'local_id': 'LINCS-2021', 'persistent_id': 'https://clue.io/data/CMap2020#LINCS2020', 'creation_time': datetime(2020, 11, 20).astimezone().isoformat(), 'abbreviation': 'LINCS_2021', 'name': 'LINCS 2021 Data Release', 'description': 'The 2021 beta release of the CMap LINCS resource', } ], columns=[ 'id_namespace', 'local_id', 'persistent_id', 'creation_time', 'abbreviation', 'name', 'description' ]).to_csv('project.tsv', sep='\t', index=False) pd.DataFrame([{ 'parent_project_id_namespace': 'https://www.lincsproject.org', 'parent_project_local_id': 'LINCS', 'child_project_id_namespace': 'https://www.lincsproject.org', 'child_project_local_id': 'LINCS-2021', }], columns=[ 'parent_project_id_namespace', 'parent_project_local_id', 'child_project_id_namespace', 'child_project_local_id' ]).to_csv('project_in_project.tsv', sep='\t', index=False) -
Determine the relationships between files and their associated samples or biological subjects, as well as all relevant assay types, analysis methods, data types, file formats, etc. Also identify all appropriate ontological mappings, if any, corresponding to each value from above.
- The LINCS DCC includes internal drug, gene, and cell line identifiers, which were mapped to PubChem, Ensembl, and Disease Ontology/UBERON manually ahead of time, but other DCCs may already make use of the CFDE-supported ontologies.
- For instance, the LINCS signature
L1000_LINCS_DCIC_ABY001_A375_XH_A16_lapatinib_10uM.tsv.gzcomes from the biosampleABY001_A375_XH_A16_lapatinib_10uM(in this case an experimental condition) and the subject cell lineA375; has a data type of expression matrix (data:0928); is stored as a TSV file format (format:3475) with GZIP compression format (format:3989); and has a MIME type oftext/tab-separated-values. - The
ABY001_A375_XH_A16_lapatinib_10uMbiosample was obtained via the L1000 sequencing assay type (OBI:0002965); comes from a cell line derived from the skin (UBERON:0002097); and was treated with the compound lapatinib (CID:208908).
-
Either manually or programmatically, generate each data table, starting with the core entity tables (
file,biosample,subject). This step will depend entirely on the format of a DCC's existing metadata and ontology mapping tables. -
Generate the inter-entity linkage association tables (
file_describes_biosample,file_describes_subject,biosample_from_subject).- In the LINCS tutorial, since the filenames come directly from the biosamples, once the
fileandbiosampletables have both been built,file_describes_biosamplecan be generated.
fdb = file_df[['id_namespace', 'local_id']].copy() fdb = fdb.rename( columns={ 'id_namespace': 'file_id_namespace', 'local_id': 'file_local_id' } ) fdb['biosample_id_namespace'] = fdb['file_id_namespace'] fdb['biosample_local_id'] = fdb['file_local_id'].apply(file_2_biosample_map_function) - In the LINCS tutorial, since the filenames come directly from the biosamples, once the
-
Assign files, biosamples, and subjects to any collections, if applicable, using the
file_in_collection,subject_in_collection, andbiosample_in_collectiontables.- Collections are optional, and can represent files from the same publications or other logical groupings outside of funding.
-
Continue with submission guide:
# finish preparing your package by resolving iris cfde-c2m2 prepare # verify integrity of your package and address any errors cfde-c2m2 validate # zip the necessary files for a bare minimum package cfde-c2m2 package -
The
C2M2_datapackage.zipfile can now be uploaded to the CFDE Workbench Metadata and Data Submission System.
Other Links
- The original C2M2 documentation from the CFDE Coordination Center contains more details on the concepts discussed here.
- The most up-to-date C2M2 JSON schema (NOTE: cfde-cli package downloads this for you automatically)
- The most up-to-date C2M2 ontology preparation script and files (NOTE: cfde-cli package downloads this for you automatically)