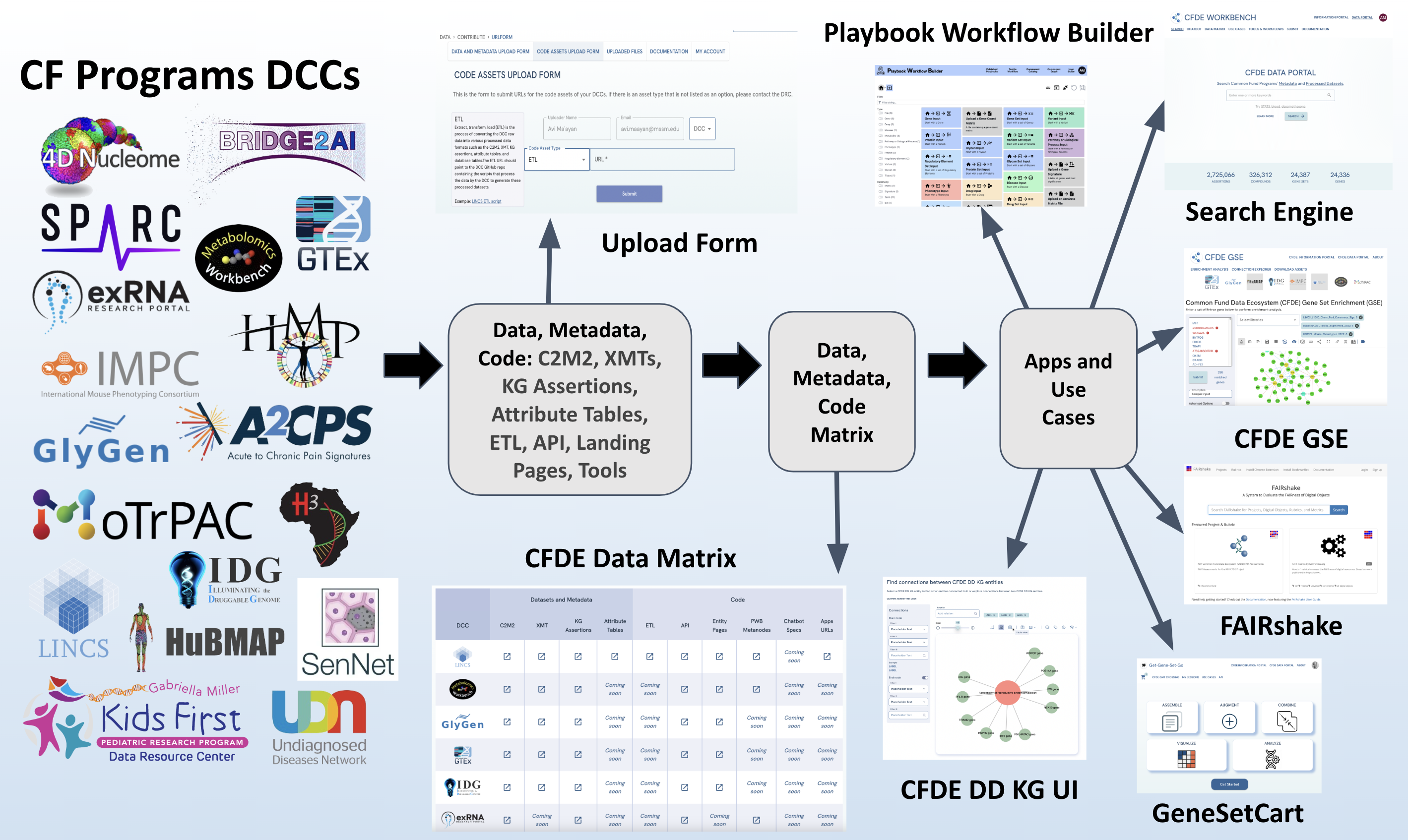
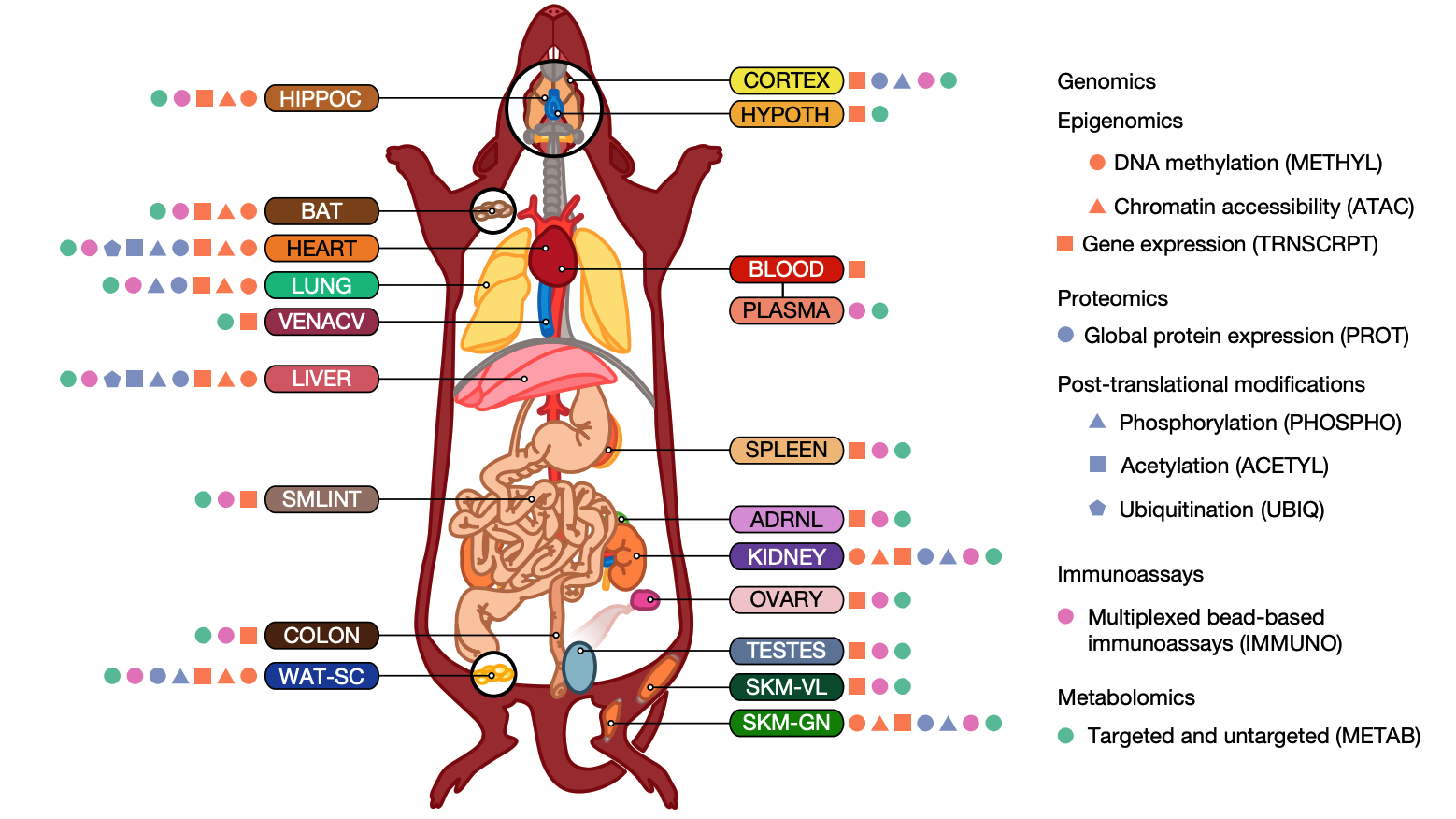
CFDE Associated and Common Fund Programs’ Landmark Publications
Contribution of Circulating Host and Microbial Tryptophan Metabolites Toward Ah Receptor Activation.
@CFDE Workbench 2026
The CFDE Workbench is actively being developed and maintained by the CFDE Data Resource Center (DRC).The DRC is funded by OT2OD036435 from the Common Fund at the National Institutes of Health.
The CFDE Workbench is actively being developed and maintained by the CFDE Data Resource Center (DRC).The DRC is funded by OT2OD036435 from the Common Fund at the National Institutes of Health.
@CFDE Workbench 2026